Data sets
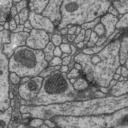
Segmented ssTEM stack of neural tissue
The challenge: use this data set to train machine learning software for the purpose of automatic segmentation of neural structures in ssTEM.
The images are representative of actual images in the real-world: there is a bit of noise; there are image registration errors; there is even a small stitching error in one section. None of these led to any difficulties in the manual labeling of each element in the image stack by an expert human neuroanatomist. A software application that aims at removing or reducing human operation must be able to cope with all these issues.
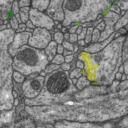
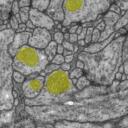
How does the data look like: each labeled object has a unique id and fits into the overall datastructure of the data set. For example, each mitochondria is represented by a unique Arealist object, containing a list of labeled areas, one per section.
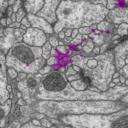
All membranes have been highlighted as one unique object. All neurites (and glia) have been highlighted each as its own independent object, delimited by membrane and non-overlapping with membrane and with each other.
On the other hand, mitochondria, noise and synapses overlap with membranes, neurites and glia; hence, they are offered as independent tif stacks.
Files:
- Seg.tar.bz2: TrakEM2 project XML file and TEM images.
- tifs.tar.bz2: All label stacks as multitif files, containing non-overlapping segmentations.
Contents:
License:
You are free to use this data set for the purpose of generating or testing non-commercial image segmentation software. If any scientific publications derive from the usage of this data set, you must cite TrakEM2 and the following publication:
Cardona A, Saalfeld S, Preibisch S, Schmid B, Cheng A, Pulokas J, Tomancak P, Hartenstein V. 2010. An Integrated Micro- and Macroarchitectural Analysis of the Drosophila Brain by Computer-Assisted Serial Section Electron Microscopy. PLoS Biol 8(10): e1000502. doi:10.1371/journal.pbio.1000502.
Contact:
Albert CardonaUpdate 2010-11-01
Exported segmentations for mitochondria have been updated. Some traced mitochondria were missing in the exported tif file with mitochondria labels. Also closed a few tiny holes in some mitochondria. Thanks to Jan Funke for spotting the missing mitochondria.
Update 2010-02-26
Exported segmentations in tif files have been updated. A TrakEM2 error, on occasions, prevented some areas from being exported as labels in the tif stack.
Thanks to Ignacio Arganda-Carreras for spotting this problem.